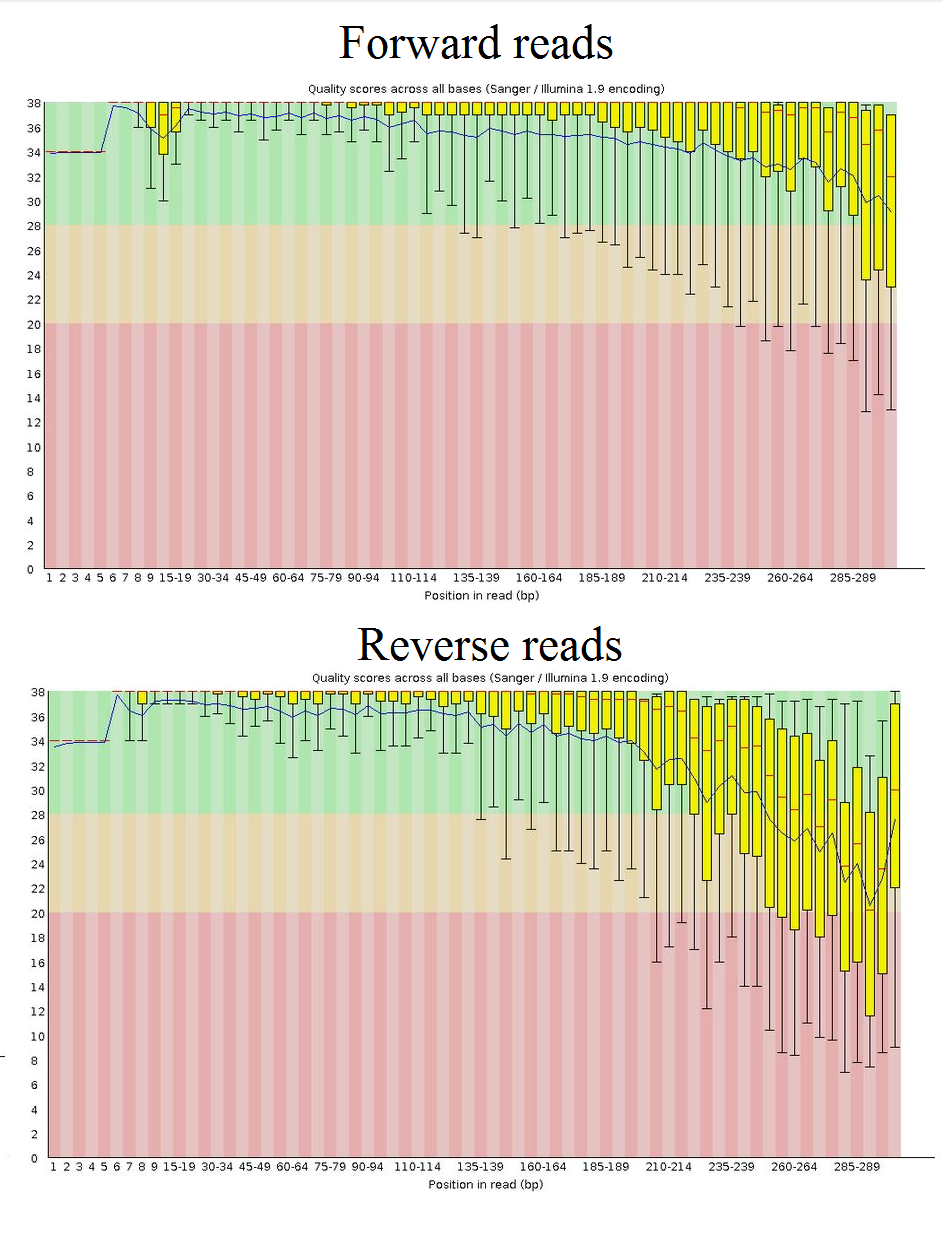
paired end sequencing read length
Refer to the How many cycles of. Leaving it at 25bp and having read length the splicing mapping is basically worthless.
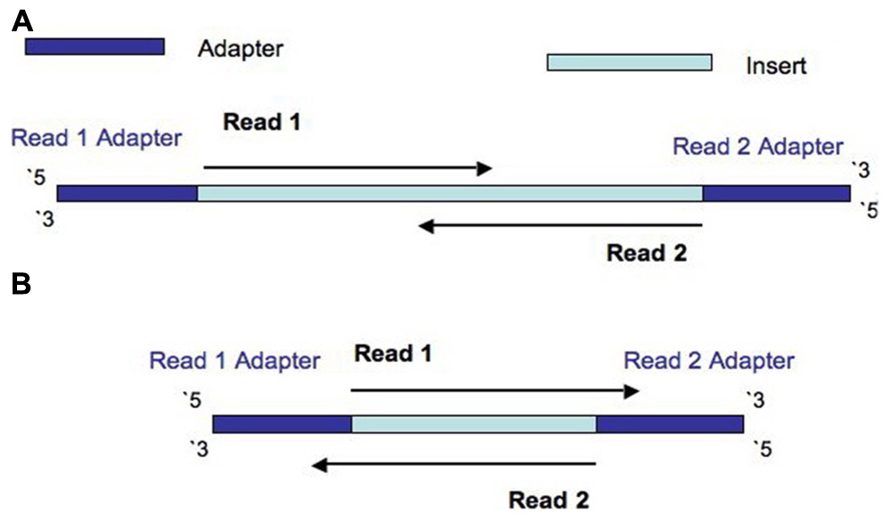
Frontiers Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
We use an Illumina MiniSeq for our short-read sequencing runs.

. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first generation sequencing. We have previously shown how different enrichment methods perform with respect to covered regions underrepresented regions and sequencing efficiency. The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phi x-musigmaxLp0 where mu is the mean.
B no overlap between the paired-end. If you only have one fragment worth ie. A Short overlap between the paired-end reads.
My paired-end read sequencing data are 50 base-pairs on each end nearly all exact with about 300bp unsequenced inner distance between the 50bp ends. You can set that 25bp to be 20 or. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the.
The library prep protocols are designed to. An analysis by Whiteford et al. To ensure the highest level of quality Illumina supports reads up to a certain length depending on the sequencing platform and SBS kit version.
The pooled indexed libraries were loaded into the MiSeq Reagent kit V2 300 cycles Illumina and sequenced on the MiSeq platform Illumina in paired-end mode with a read. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase.
On sequencing using unpaired reads shows that ultra-short reads theoretically allow whole genome re-sequencing and de novo assembly of only small. The paired-end short read lengths are always 2 x 150bp 300bp. Three possible scenarios for paired-end read lengths and target DNA fragment lengths.
Direct RNA sequencing also brings the benefit of accurate. In this Tech Note we focus on. The longest transcript sequenced by nanopore sequencing currently stands at over 20 kb in length.

Five Approaches To Detect Cnvs From Ngs Short Reads A Paired End Download Scientific Diagram

Schematic Representation Of Paired End Sequencing For Very Short Download Scientific Diagram
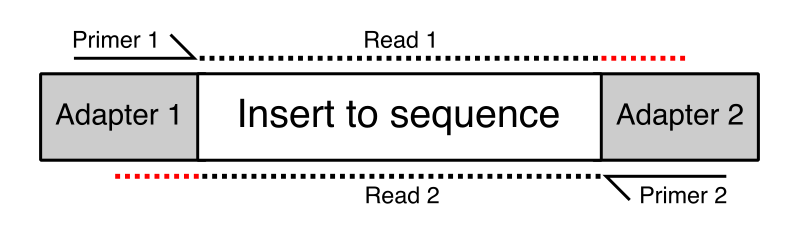
Qc Fail Sequencing Read Through Adapters Can Appear At The Ends Of Sequencing Reads

Stacks 2 Analytical Methods For Paired End Sequencing Improve Radseq Based Population Genomics Rochette 2019 Molecular Ecology Wiley Online Library
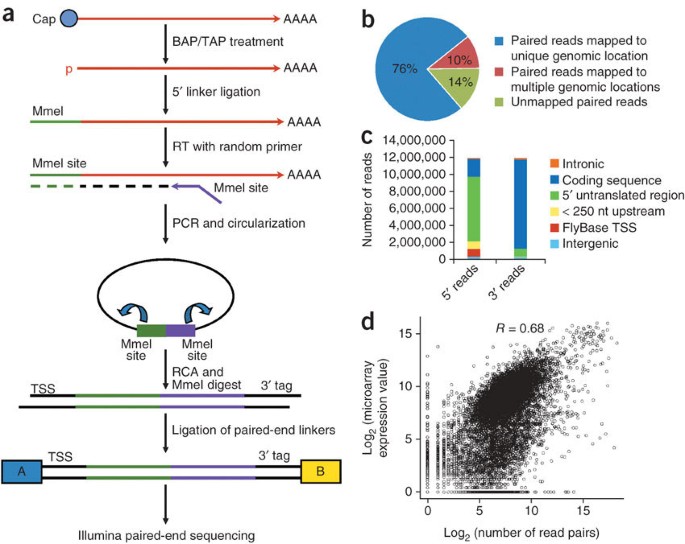
A Paired End Sequencing Strategy To Map The Complex Landscape Of Transcription Initiation Nature Methods

Atac Seq Guidelines Harvard Fas Informatics

Interpreting Color By Insert Size Integrative Genomics Viewer

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download High Resolution Scientific Diagram

Casava Files Of Paired End Reads General Discussion Qiime 2 Forum
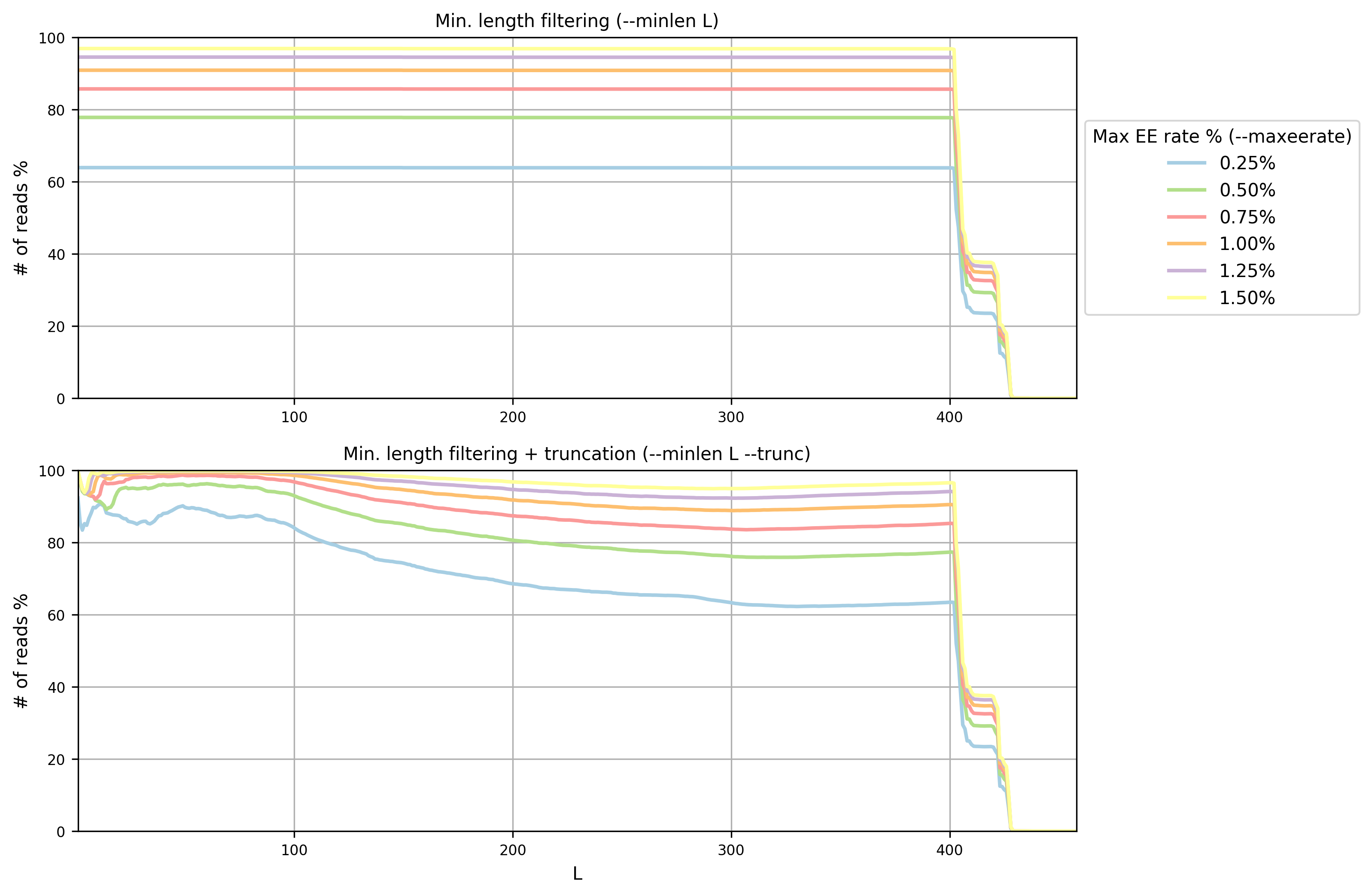
Paired End Sequencing 97 Otu Micca 1 7 0 Documentation

Dnbseq G50 Gets Full Performance Upgrade Mgi Leading Life Science Innovation
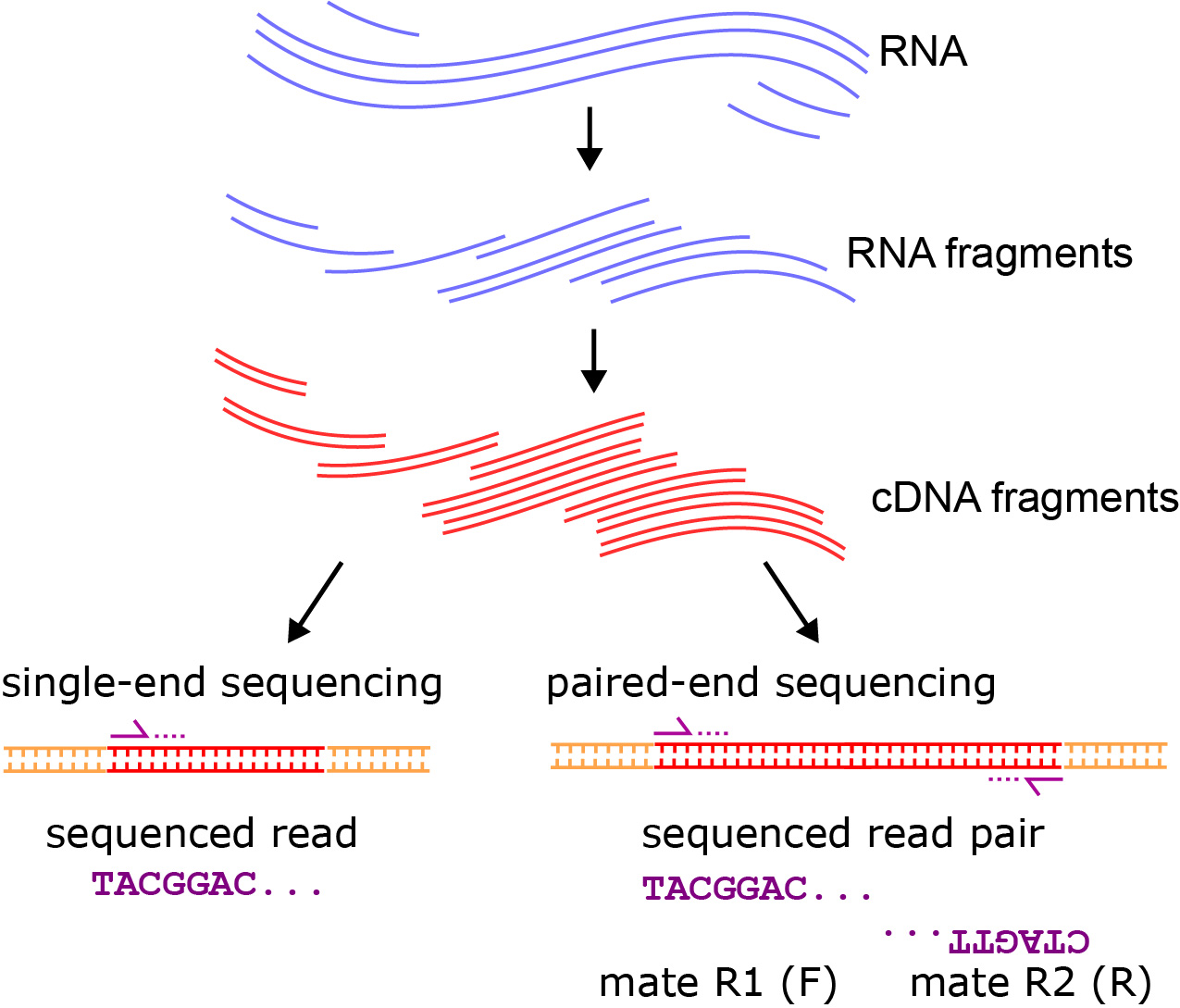
Chapter 6 Transcriptomics Applied Bioinformatics
What Is Paired End 150 Omega Bioservices

Paired End Vs Single End Sequencing Reads Youtube

Compbio 020 Reads Fragments And Inserts What You Need To Know For Understanding Your Sequencing Data Bad Grammar Good Syntax
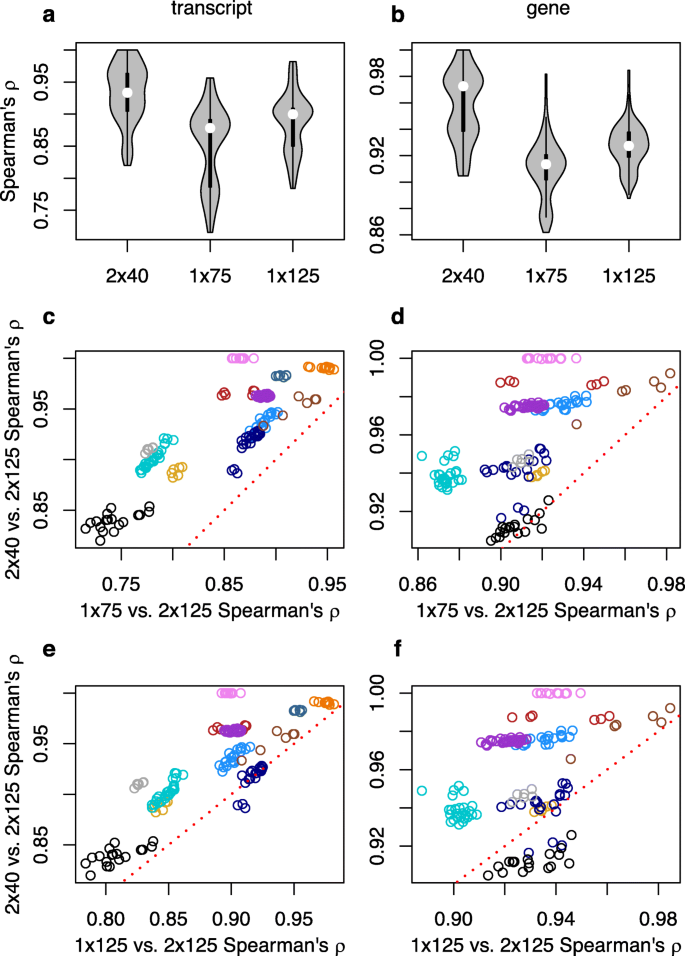
Short Paired End Reads Trump Long Single End Reads For Expression Analysis Bmc Bioinformatics Full Text